Creates a list that explains the beta values, and their corrections, of the ensemble IBLM model
Arguments
- iblm_model
An object of class 'iblm'. This should be output by `train_iblm_xgb()`
- data
Data frame.
If you have used `split_into_train_validate_test()` this will be the "test" portion of your data.
- migrate_reference_to_bias
Logical, migrate the beta corrections to the bias for reference levels? This applied to categorical vars only. It is recommended to leave this setting on TRUE
Value
A list containing:
- beta_corrected_scatter
Function to create scatter plots showing SHAP corrections vs variable values (see
beta_corrected_scatter)- beta_corrected_density
Function to create density plots of SHAP corrections for variables (see
beta_corrected_density)- bias_density
Function to create density plots of SHAP corrections migrated to bias (see
bias_density)- overall_correction
Function to show global correction distributions (see
overall_correction)- shap
Dataframe showing raw SHAP values of data records
- beta_corrections
Dataframe showing beta corrections (in wide/one-hot format) of data records
- data_beta_coeff
Dataframe showing beta coefficients of data records
Details
The following outputs are functions that can be called to create plots:
beta_corrected_scatter
beta_corrected_density
bias_density
overall_correction
For each of these, the key data arguments (e.g. data, shap, iblm_model) are already populated by `explain_iblm()`. When calling these functions output from `explain_iblm()` only key settings like variable names, colours...etc need populating.
Examples
df_list <- freMTPLmini |> split_into_train_validate_test(seed = 9000)
iblm_model <- train_iblm_xgb(
df_list,
response_var = "ClaimRate",
family = "poisson"
)
ex <- explain_iblm(iblm_model, df_list$test)
# the output contains functions that can be called to visualise iblm
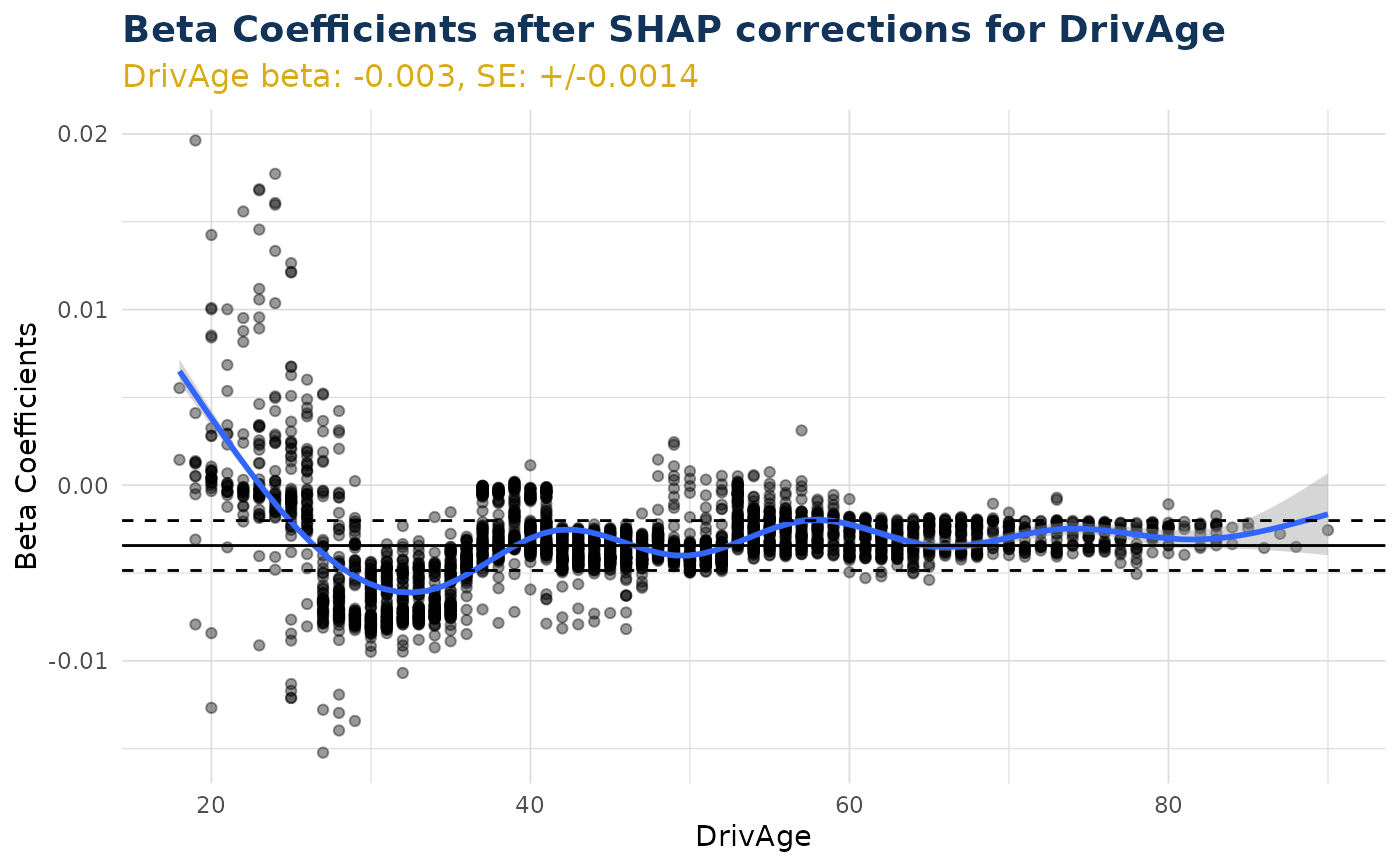
ex$beta_corrected_scatter("DrivAge")
#> `geom_smooth()` using method = 'gam' and formula = 'y ~ s(x, bs = "cs")'
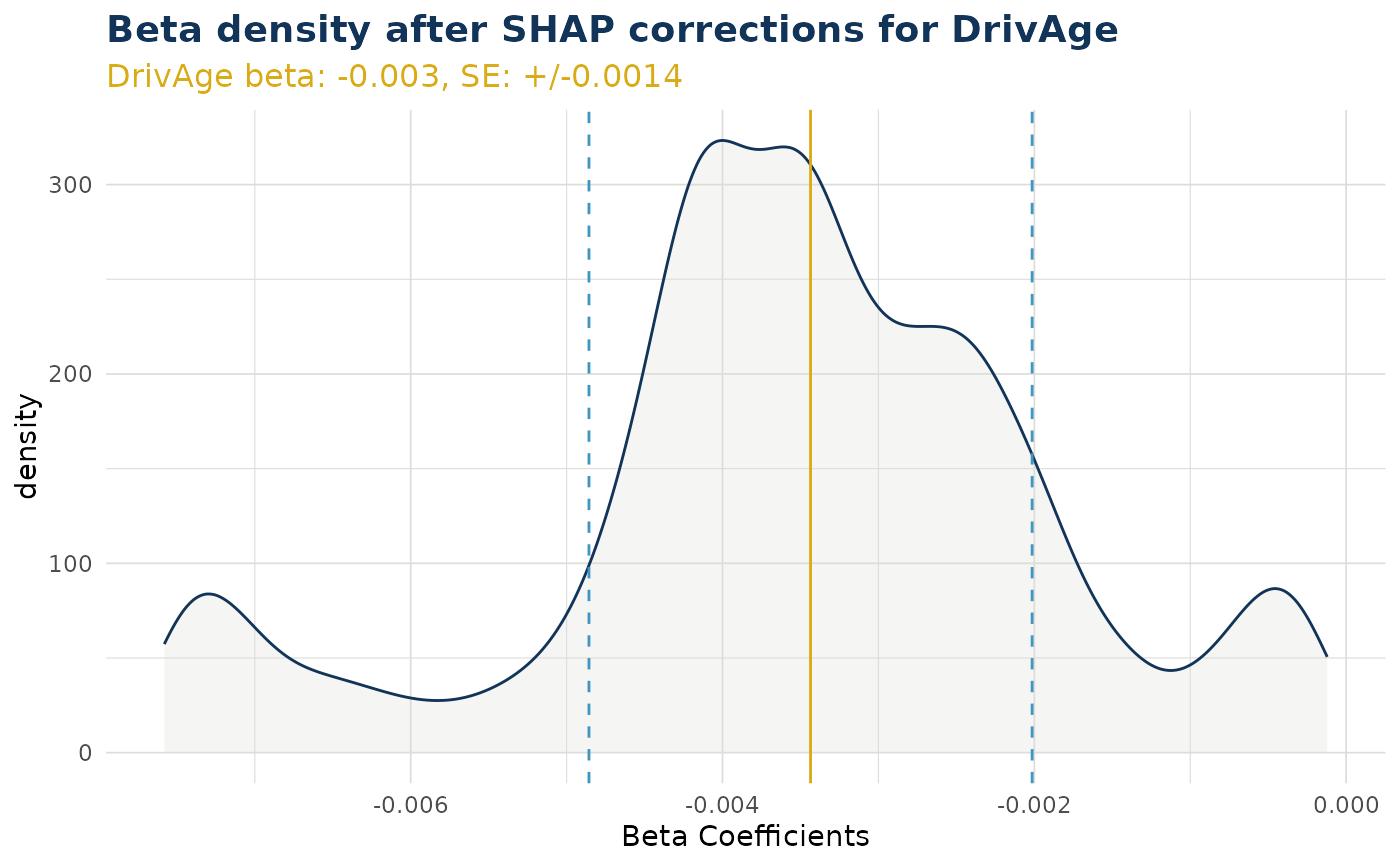
 ex$beta_corrected_density("DrivAge")
ex$beta_corrected_density("DrivAge")
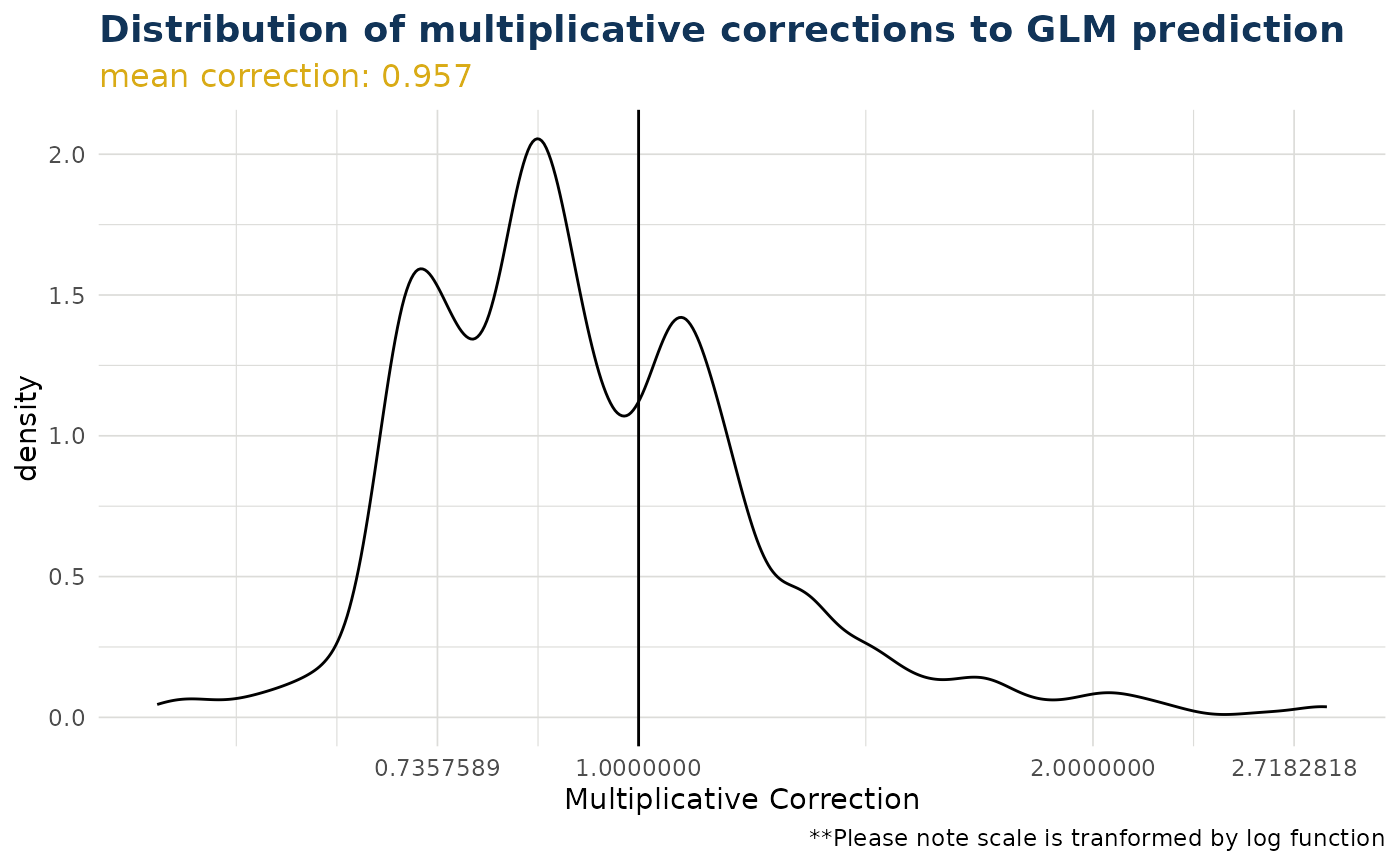
 ex$overall_correction()
ex$overall_correction()
 ex$bias_density()
#> $bias_correction_var
ex$bias_density()
#> $bias_correction_var
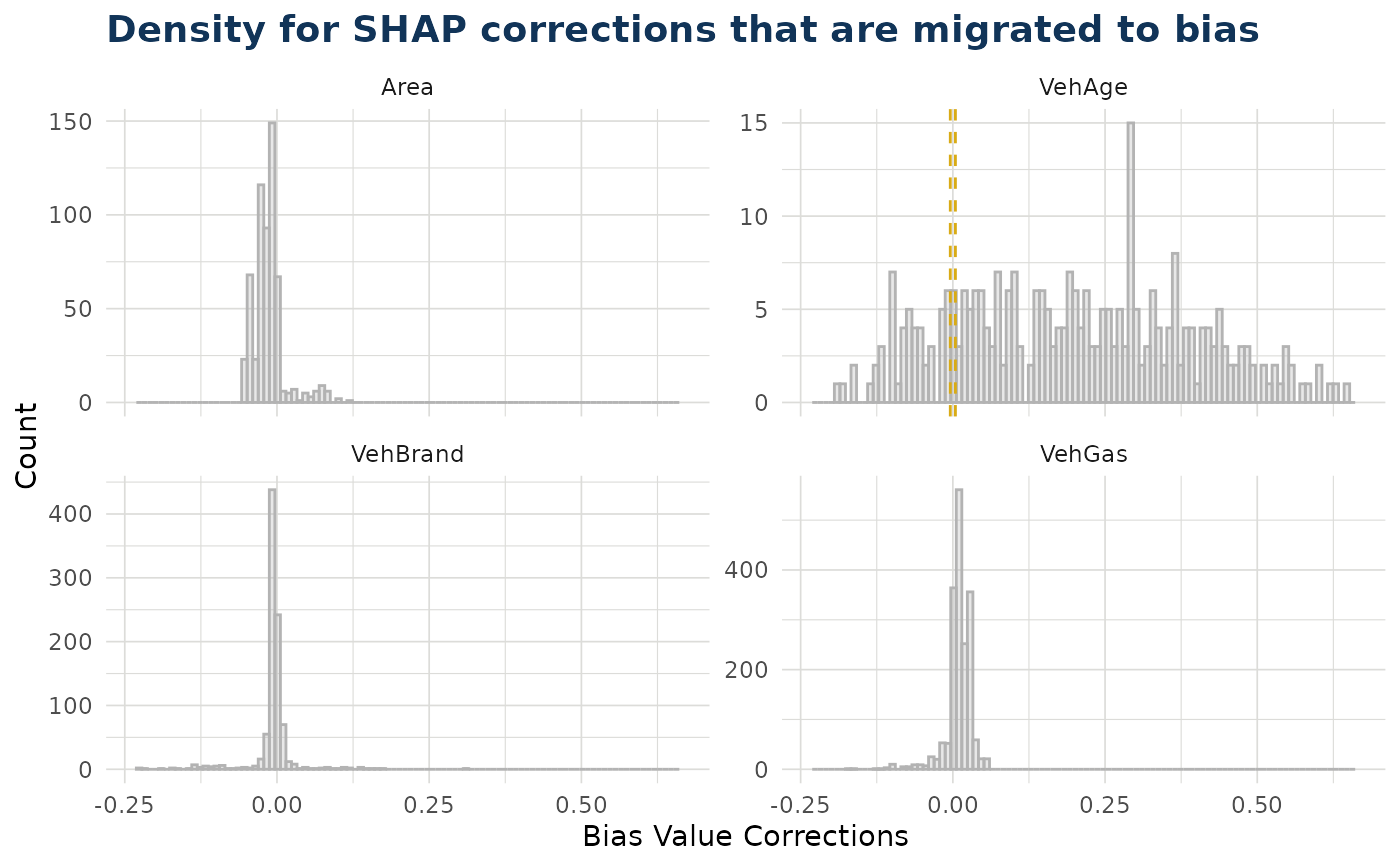 #>
#> $bias_correction_total
#>
#> $bias_correction_total
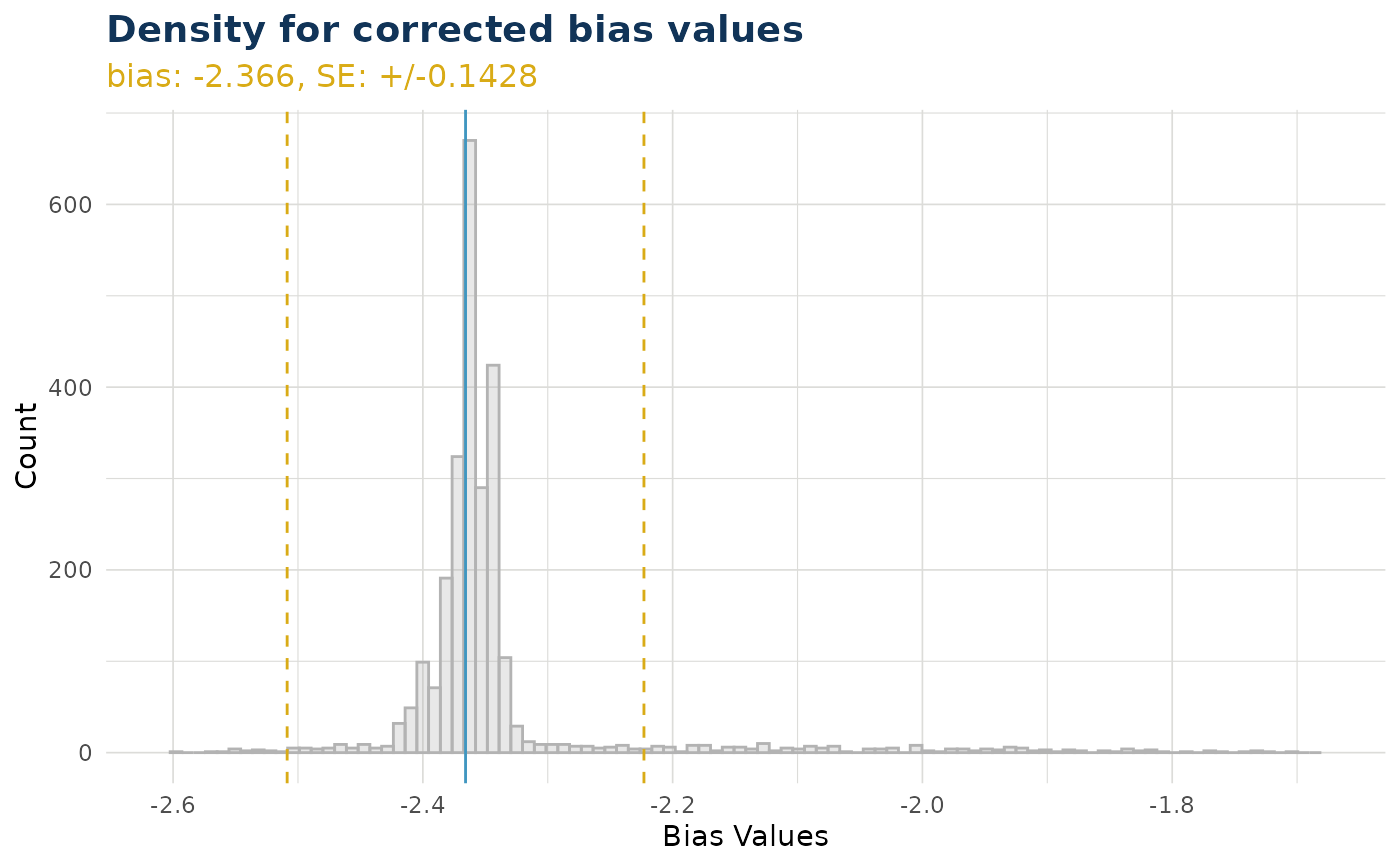 #>
# the output contains also dataframes
ex$shap |> dplyr::glimpse()
#> Rows: 3,764
#> Columns: 8
#> $ Area <dbl> -0.017667860, 0.016070237, 0.003410609, -0.009702625, -0.02…
#> $ VehPower <dbl> -9.513386e-03, -1.709264e-02, 1.250970e-02, 5.950155e-03, -…
#> $ VehAge <dbl> -0.1181081235, -0.1903213263, 0.0632225871, 0.0886134803, 0…
#> $ DrivAge <dbl> -0.0106765414, 0.0178347062, -0.0178089067, 0.1007688642, -…
#> $ BonusMalus <dbl> -0.009905483, 0.035999734, -0.025095381, 0.007291305, -0.03…
#> $ VehBrand <dbl> -6.775799e-02, -7.489327e-02, -2.930630e-02, 4.647116e-02, …
#> $ VehGas <dbl> 0.0016007025, 0.0040426101, 0.0229403824, -0.0065398184, 0.…
#> $ BIAS <dbl> -0.03920801, -0.03920801, -0.03920801, -0.03920801, -0.0392…
ex$beta_corrections |> dplyr::glimpse()
#> Rows: 3,764
#> Columns: 24
#> $ bias <dbl> -0.037607307, -0.093988436, -0.016267627, -0.039208010, …
#> $ VehPower <dbl> -8.648532e-04, -3.418528e-03, 2.084949e-03, 1.190031e-03…
#> $ VehAge <dbl> -1.181081e-01, -1.903213e-01, 7.902823e-03, 6.329534e-03…
#> $ DrivAge <dbl> -1.840783e-04, 7.133882e-04, -4.240216e-04, 1.399568e-03…
#> $ BonusMalus <dbl> -1.981097e-04, 3.789446e-04, -5.019076e-04, 1.458261e-04…
#> $ AreaA <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
#> $ AreaB <dbl> 0.00000000, 0.00000000, 0.00000000, 0.00000000, 0.000000…
#> $ AreaC <dbl> 0.000000000, 0.000000000, 0.000000000, -0.009702625, -0.…
#> $ AreaD <dbl> 0.000000000, 0.000000000, 0.003410609, 0.000000000, 0.00…
#> $ AreaE <dbl> -0.017667860, 0.000000000, 0.000000000, 0.000000000, 0.0…
#> $ AreaF <dbl> 0.0000000, 0.0000000, 0.0000000, 0.0000000, 0.0000000, 0…
#> $ VehBrandB1 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
#> $ VehBrandB10 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
#> $ VehBrandB11 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
#> $ VehBrandB12 <dbl> -0.06775799, 0.00000000, 0.00000000, 0.00000000, 0.00000…
#> $ VehBrandB13 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
#> $ VehBrandB14 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
#> $ VehBrandB2 <dbl> 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, …
#> $ VehBrandB3 <dbl> 0.0000000, 0.0000000, -0.0293063, 0.0000000, 0.0000000, …
#> $ VehBrandB4 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
#> $ VehBrandB5 <dbl> 0.00000000, 0.00000000, 0.00000000, 0.04647116, 0.000000…
#> $ VehBrandB6 <dbl> 0.000000000, 0.000000000, 0.000000000, 0.000000000, 0.00…
#> $ VehGasDiesel <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
#> $ VehGasRegular <dbl> 0.000000000, 0.000000000, 0.000000000, -0.006539818, 0.0…
ex$data_beta_coeff |> dplyr::glimpse()
#> Rows: 3,764
#> Columns: 8
#> $ bias <dbl> -2.403428, -2.459809, -2.382088, -2.405029, -2.381321, -2.4…
#> $ Area <dbl> -0.021937324, 0.000000000, 0.069380839, -0.147578696, -0.16…
#> $ VehPower <dbl> -0.006312828, -0.008866502, -0.003363025, -0.004257944, -0.…
#> $ VehAge <dbl> -0.15942935, -0.23164256, -0.03341841, -0.03499170, -0.0368…
#> $ DrivAge <dbl> -0.003619471, -0.002722004, -0.003859414, -0.002035825, -0.…
#> $ BonusMalus <dbl> 0.01173046, 0.01230752, 0.01142666, 0.01207440, 0.01129568,…
#> $ VehBrand <dbl> 0.647894859, 0.000000000, -0.001259444, 0.767806208, 0.0182…
#> $ VehGas <dbl> 0.0000000, 0.0000000, 0.0000000, 0.2588874, 0.0000000, 0.25…
#>
# the output contains also dataframes
ex$shap |> dplyr::glimpse()
#> Rows: 3,764
#> Columns: 8
#> $ Area <dbl> -0.017667860, 0.016070237, 0.003410609, -0.009702625, -0.02…
#> $ VehPower <dbl> -9.513386e-03, -1.709264e-02, 1.250970e-02, 5.950155e-03, -…
#> $ VehAge <dbl> -0.1181081235, -0.1903213263, 0.0632225871, 0.0886134803, 0…
#> $ DrivAge <dbl> -0.0106765414, 0.0178347062, -0.0178089067, 0.1007688642, -…
#> $ BonusMalus <dbl> -0.009905483, 0.035999734, -0.025095381, 0.007291305, -0.03…
#> $ VehBrand <dbl> -6.775799e-02, -7.489327e-02, -2.930630e-02, 4.647116e-02, …
#> $ VehGas <dbl> 0.0016007025, 0.0040426101, 0.0229403824, -0.0065398184, 0.…
#> $ BIAS <dbl> -0.03920801, -0.03920801, -0.03920801, -0.03920801, -0.0392…
ex$beta_corrections |> dplyr::glimpse()
#> Rows: 3,764
#> Columns: 24
#> $ bias <dbl> -0.037607307, -0.093988436, -0.016267627, -0.039208010, …
#> $ VehPower <dbl> -8.648532e-04, -3.418528e-03, 2.084949e-03, 1.190031e-03…
#> $ VehAge <dbl> -1.181081e-01, -1.903213e-01, 7.902823e-03, 6.329534e-03…
#> $ DrivAge <dbl> -1.840783e-04, 7.133882e-04, -4.240216e-04, 1.399568e-03…
#> $ BonusMalus <dbl> -1.981097e-04, 3.789446e-04, -5.019076e-04, 1.458261e-04…
#> $ AreaA <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
#> $ AreaB <dbl> 0.00000000, 0.00000000, 0.00000000, 0.00000000, 0.000000…
#> $ AreaC <dbl> 0.000000000, 0.000000000, 0.000000000, -0.009702625, -0.…
#> $ AreaD <dbl> 0.000000000, 0.000000000, 0.003410609, 0.000000000, 0.00…
#> $ AreaE <dbl> -0.017667860, 0.000000000, 0.000000000, 0.000000000, 0.0…
#> $ AreaF <dbl> 0.0000000, 0.0000000, 0.0000000, 0.0000000, 0.0000000, 0…
#> $ VehBrandB1 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
#> $ VehBrandB10 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
#> $ VehBrandB11 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
#> $ VehBrandB12 <dbl> -0.06775799, 0.00000000, 0.00000000, 0.00000000, 0.00000…
#> $ VehBrandB13 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
#> $ VehBrandB14 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
#> $ VehBrandB2 <dbl> 0.000000e+00, 0.000000e+00, 0.000000e+00, 0.000000e+00, …
#> $ VehBrandB3 <dbl> 0.0000000, 0.0000000, -0.0293063, 0.0000000, 0.0000000, …
#> $ VehBrandB4 <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
#> $ VehBrandB5 <dbl> 0.00000000, 0.00000000, 0.00000000, 0.04647116, 0.000000…
#> $ VehBrandB6 <dbl> 0.000000000, 0.000000000, 0.000000000, 0.000000000, 0.00…
#> $ VehGasDiesel <dbl> 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,…
#> $ VehGasRegular <dbl> 0.000000000, 0.000000000, 0.000000000, -0.006539818, 0.0…
ex$data_beta_coeff |> dplyr::glimpse()
#> Rows: 3,764
#> Columns: 8
#> $ bias <dbl> -2.403428, -2.459809, -2.382088, -2.405029, -2.381321, -2.4…
#> $ Area <dbl> -0.021937324, 0.000000000, 0.069380839, -0.147578696, -0.16…
#> $ VehPower <dbl> -0.006312828, -0.008866502, -0.003363025, -0.004257944, -0.…
#> $ VehAge <dbl> -0.15942935, -0.23164256, -0.03341841, -0.03499170, -0.0368…
#> $ DrivAge <dbl> -0.003619471, -0.002722004, -0.003859414, -0.002035825, -0.…
#> $ BonusMalus <dbl> 0.01173046, 0.01230752, 0.01142666, 0.01207440, 0.01129568,…
#> $ VehBrand <dbl> 0.647894859, 0.000000000, -0.001259444, 0.767806208, 0.0182…
#> $ VehGas <dbl> 0.0000000, 0.0000000, 0.0000000, 0.2588874, 0.0000000, 0.25…
